
CHEBI:52742
| Name | D-ribofuranose 5-phosphate | 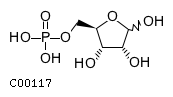 |
| Synonyms | 05-o-phosphono-d-ribose; D-ribose 5-phosphate; D-ribose 5-phosphate; Ribose 5-phosphate; | |
| Definition | The furanose form of D-ribose 5-phosphate. | |
| Molecular Weight (Exact mass) | 230.1098 (230.0192) | |
| Molecular Formula | C5H11O8P | |
| SMILES | OC1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H]1O | |
| InChI | InChI=1S/C5H11O8P/c6-3-2(1-12-14(9,10)11)13-5(8)4(3)7/h2-8H,1H2,(H2,9,10,11)/t2-,3-,4-,5?/m1/s1 | |
| InChI Key | KTVPXOYAKDPRHY-SOOFDHNKSA-N | |
| Crosslinking annotations | KEGG:C00117 | 3DMET:B04635 | CAS:4300-28-1 | ChEBI:17797 | ChEBI:52742 | ChEMBL:CHEMBL1235722 | ChEMBL:CHEMBL605020 | KNApSAcK:C00007473 | NIKKAJI:J205.693C | PDB-CCD:HSX | PDB-CCD:RP5 | PubChem:3417 |
| Pathway ID | Pathway Name | Pathway Description (KEGG) |
| map00030 | Pentose phosphate pathway | The pentose phosphate pathway is a process of glucose turnover that produces NADPH as reducing equivalents and pentoses as essential parts of nucleotides. There are two different phases in the pathway. One is irreversible oxidative phase in which glucose-6P is converted to ribulose-5P by oxidative decarboxylation, and NADPH is generated [MD:M00006]. The other is reversible non-oxidative phase in which phosphorylated sugars are interconverted to generate xylulose-5P, ribulose-5P, and ribose-5P [MD:M00007]. Phosphoribosyl pyrophosphate (PRPP) formed from ribose-5P [MD:M00005] is an activated compound used in the biosynthesis of histidine and purine/pyrimidine nucleotides. This pathway map also shows the Entner-Doudoroff pathway where 6-P-gluconate is dehydrated and then cleaved into pyruvate and glyceraldehyde-3P [MD:M00008]. |
| map00230 | Purine metabolism | NA |
| map00440 | Phosphonate and phosphinate metabolism | Natural products containing carbon-phosphorous bonds, so-called C-P compounds, are derivatives of phosphonate and phosphinate with substitution of alkyl group for hydrogen of phosphorus-hydrogen bonds. C-P compounds have been found in many organisms, but only protists and bacteria, mostly Actinobacteria, have biosynthetic capacity. A common reaction in the biosynthetic pathway is C-P bond forming reaction from phosphoenolpyruvate (PEP) to phosphonopyruvate (PnPy) catalyzed by PEP phosphomutase. 2-Aminoethylphosphonate (AEP) is the most abundant C-P compound in the natural world. AEP derivatives include phosphonoprotein, phosphonoglycan, and phosphonolipid. Other known C-P compounds are bioactive substances used in medicine (antibiotics) and agriculture (herbicide) such as fosfomycin, FR-33289, rhizocticin, and bialaphos. |
| map00710 | Carbon fixation in photosynthetic organisms | NA |
| map01100 | Metabolic pathways | NA |
| map01110 | Biosynthesis of secondary metabolites | NA |
| map01120 | Microbial metabolism in diverse environments | NA |
| map01130 | Biosynthesis of antibiotics | NA |
| map01200 | Carbon metabolism | Carbon metabolism is the most basic aspect of life. This map presents an overall view of central carbon metabolism, where the number of carbons is shown for each compound denoted by a circle, excluding a cofactor (CoA, CoM, THF, or THMPT) that is replaced by an asterisk. The map contains carbon utilization pathways of glycolysis (map00010), pentose phosphate pathway (map00030), and citrate cycle (map00020), and six known carbon fixation pathways (map00710 and map00720) as well as some pathways of methane metabolism (map00680). The six carbon fixation pathways are: (1) reductive pentose phosphate cycle (Calvin cycle) in plants and cyanobacteria that perform oxygenic photosynthesis, (2) reductive citrate cycle in photosynthetic green sulfur bacteria and some chemolithoautotrophs, (3) 3-hydroxypropionate bi-cycle in photosynthetic green nonsulfur bacteria, two variants of 4-hydroxybutyrate pathways in Crenarchaeota called (4) hydroxypropionate-hydroxybutyrate cycle and (5) dicarboxylate-hydroxybutyrate cycle, and (6) reductive acetyl-CoA pathway in methanogenic bacteria. |
| map01230 | Biosynthesis of amino acids | This map presents a modular architecture of the biosynthesis pathways of twenty amino acids, which may be viewed as consisting of the core part and its extensions. The core part is the KEGG module for conversion of three-carbon compounds from glyceraldehyde-3P to pyruvate [MD:M00002], together with the pathways around serine and glycine. This KEGG module is the most conserved one in the KEGG MODULE database and is found in almost all the completely sequenced genomes. The extensions are the pathways containing the reaction modules RM001, RM033, RM032, and RM002 for biosynthesis of branched-chain amino acids (left) and basic amino acids (bottom), and the pathways for biosynthesis of histidine and aromatic amino acids (top right). It is interesting to note that the so-called essential amino acids that cannot be synthesized in human and other organisms generally appear in these extensions. Furthermore, the bottom extension of basic amino acids appears to be most divergent containing multiple pathways for lysine biosynthesis and multiple gene sets for arginine biosynthesis. |
| map04918 | Thyroid hormone synthesis | Thyroid hormones triiodothyronine (T3) and thyroxine (T4) are essential for normal development, growth and metabolic homeostasis in all vertebrates, and synthesized in the thyroid gland. The functional unit of the thyroid gland is the follicle, delimited by a monolayer of thyrocytes. Polarized thyrocytes surround the follicular lumen; with their basal and apical surfaces facing the bloodstream and the lumen, respectively. To synthesize thyroid hormones, thyrocytes take up iodide at their basal side and concentrate it into the lumen. They also secrete in this lumen the specialized protein thyroglobulin (TG) which serves as a store for the hormones. In the follicular lumen oxidation of iodine, iodination of tyrosines (MIT, 3-monoiodotyrosine; DIT, 3,5-diiodotyrosine) and coupling of iodotyrosines takes place on tyrosine residues in TG, resulting in T3 and T4 synthesis. Iodinated TG is resorbed through the apical membrane and degraded to form T3/T4 in lysosomes; the T3/T4 is then secreted through the basal membrane. |

